Last week, IBM together with health insurer WellPoint Inc., and New York’s Memorial Sloan-Kettering Cancer Center announced the commercialization of Watson (the supercomputer which beat human champions in "Jeopardy!" on February 16, 2011) for question answering (QA) in the clinical domain. The following are some interesting facts released by IBM as part of this announcement:
- The supercomputer has ingested 1,500 lung cancer cases from Sloan-Kettering records, plus 2 million pages of text from journals, textbooks and treatment guidelines. This is what I called Big Data in medicine.
- In 2012, Watson became 240 percent faster and 75 percent smaller so it can run on a single server. No surprise here and I expect this trend to continue.
The following YouTube video entitled Oncology Diagnosis and Treatment explains how IBM envisions using Watson for Clinical Question Answering (CQA):
The User Experience in the Watson Demo
- Clinical questions can be posed in natural language (spoken or typed in by the clinician using a keyboard).
- The sources used for answering clinical questions include both structured (EMR databases) and unstructured information (journal articles, clinical guidelines, etc.).
- Personalized medicine: the proposed interventions are driven by the data in the patient's medical record and the system can prompt the clinician for additional information on the patient if necessary. The displayed evidence and recommendations are updated to reflect changes in the patient's clinical data.
- Human Factors: the clinician is always in the loop. She can ask Watson how it arrives at a specific care recommendation and can even remove a specific evidence (if deemed irrelevant or not appropriate).
- The use of confidence scoring and evidence highlighting.
- Patient-centeredness and shared decision making: the treatment plans take into account the values, goals, and wishes of the patient (patient preferences). Treatment options are discussed with the patient.
- Comparative effectiveness is used to compare the benefits and harms of different interventions.
- Information is displayed using data visualization (dashboard) to help meet key performance indicators in the context of a value-based payment model.
The Science Behind Watson
The real question is how do we make intelligent health IT systems like Watson widely available to all patients. A landmark report published by the Institute of Medicine in 2001 and titled Crossing the Quality Chasm - A New Health System for the 21st Century contained the following recommendation:
Patients should receive care based on the best available scientific knowledge. Care should not vary illogically from clinician to clinician or from place to place.
For the scientifically (and Artificial Intelligence) inclined, the following are some pointers on the science behind Watson:
- The AI Behind Watson - The Technical Article
- Building Watson: An Overview of the DeepQA Project
- Unstructured Information Management (The IBM Journal of Research and Development)
- Semantic Web Technology in Watson.
The picture below represents a high level architecture of Watson (click on the image to enlarge it).
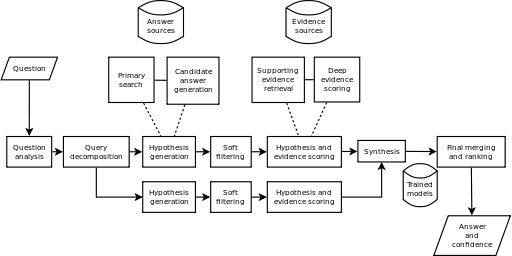
AskHermes and MiPACQ
IBM Watson is not the only effort to develop automated CQA capabilities. Some earlier CQA efforts used the PICO framework (Problem/Population, Intervention, Comparison, Outcome) to facilitate processing. More recent efforts have focused on the use of clinical questions posed in natural language.
AskHermes (Help clinicians to Extract and aRrticulate Multimedia information for answering clinical quEstionS) allows clinicians to enter questions in natural language and uses the following unstructured information sources: MEDLINE abstracts, PubMed Central full-text articles, eMedicine documents, clinical guidelines, and Wikipedia articles.
The processing pipeline in AskHermes includes the following: Question Analysis, Related Questions Extraction, Information Retrieval, Summarization and Answer Presentation. AskHermes performs question classification using MMTx (MetaMap Technology Transfer) to map keywords to UMLS concepts and semantic types. Classification is also achieved through supervised machine learning algorithms such as Support Vector Machine (SVM) and conditional random fields (CFRs). Summarization and answer presentation are based on clustering techniques.
MiPACQ (Multi-source Integrated Platform for Answering Clinical Questions) is based on Natural Language Processing (NLP) and Information Retrieval (IR) and utilizes data sources such as Electronic Medical Record (EMR) databases and online medical encyclopedia like Medpedia. MiPACQ uses a processing pipeline based on UIMA (Unstructured Information Management Architecture) and machine learning-based as well as rule-based scoring. NLP capabilities are provided by ClearTK and cTakes (clinical Text Analysis and Knowledge Extraction System).
The Road Ahead
Automated Clinical Question Answering (CQA) is really hard. However, that is the future of computing: intelligent machines we can have meaningful conversations with. CQA is a multidisciplinary field which combines disciplines like statistical computing, information retrieval, natural language processing, machine learning, rule engines, semantic web technologies, knowledge representation and reasoning, visual analytics, and massively parallel computing. There are several open source projects that provide the building blocks. Many EHR software today are glorified data entry systems. We need to move to the next level and that will require technical leadership.





No comments:
Post a Comment